Activity Analysis
Download the ActivitySequenceAnalyzer (zip file, 12.5 MB)
ActivitySequenceAnalyzer is a tool to model students' activity sequences in a Physics course as logged by the Intelligent Tutoring System Andes (VanLehn et al., 2005) and made publicly available via the PSLC DataShop (Koedinger et al., 2010).
This tool starts from the Andes data and created DMM-based models of the students' activities. In a next step, the models are serialized and processed by a clustering algorithm. The clustering process aims at the automated detection of different problem-solving dimensions and styles within student behaviour. The process can be run with different numbers of clusters and determines the most promising cluster setting by the computation of different quality metrics. The result file provides information about these metrics and the concrete content of the different clusters in order to have it further analyzed by a human expert. Please refer to (Köck & Paramythis, 2011) for more detailed information on the theoretical aspects of ActivitySequenceAnalyzer.
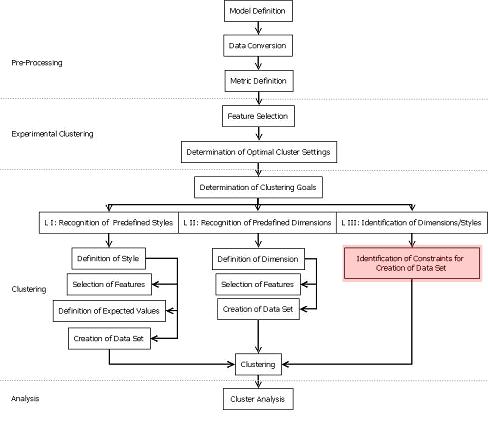
The clustering process is described in the figure below. The ActivitySequenceAnalyzer tool covers the overall process except for the "Identification of Constraints for Creation of Data Set" part.
Data
Raw data processable by ActivitySequenceAnalyzer is supposed to be in the form as exported from PSLC DataShop and contains content as can be found in the test file pslc_testdata.txt. The first line presents the types of information that are expected in the data. Data must strictly follow this scheme, different parts of information must not be switched, a tab must be used to separate the parts from each other. A line break denotes a new data instance.
The tool determines related sequences in the raw data that belong to the same student/problem combination and coverts the related data to Markov models. The models are later extended by additional statistical information like the number of attempts a student needed so solve a task. The resulting files of this step are in ARFF format and contain both attributes retrieved from the Markov models and attributes resulting from the additional statistical information. The clustering process then runs on this data and produces an output file which contains the final results for this clustering run. The attributes derived from the data and stored in the models are predefined. However, the user can chose to eliminate attributes for the clustering which is done via specific settings in the configuration file (see below).
How to use
The ActivitySequenceAnalyzer.zip file contains a build directory which again contains an executable: asa.bat. A configuration file is needed to start the process. This file must either be placed in the same directory as the .bat file and be named anaylsis.config or be provided as a parameter when the .bat is started.
The configuration file must contain the following information:
#=============================================================================
#
# ASCOLLA Activity Sequence Analyzer
# ----------------------------------
#
# Sample configuration file
#
#=============================================================================
# The path to the input file
# (mandatory)
input_file = /path/to/input/file/
# The path to the output directory
# (optional, defaults to the same directory as the input file)
output_dir = /path/to/output/directory/
# The path to the directory where intermediate files are stored
# (optional, defaults to the system's default temporary directory)
temp_dir = /path/to/temp/directory/
# Controls whether intermediate files are kept of deleted at the end
# (optional, default value is true)
keep_intermediate_files = true
# Controls whether intermediate files should be time-stamped
# (optional, defaults to true)
timestamp_files = true
# Hint mode
# (optional, default value is 0)
hint_mode = 0
# Minimum number of clusters to try
# (mandatory)
clusters.min = 2
# Maximum number of clusters to try
# (mandatory)
clusters.max = 20
# The activity model attributes to use when clustering
# (mandatory)
attributes.all = STUDENT, PROBLEM, COMPLETED, PRIOR_PROB_0, PRIOR_PROB_1, PRIOR_PROB_3, PRIOR_PROB_4, PRIOR_PROB_5, TRANS_PROB_0_0, TRANS_PROB_0_1, TRANS_PROB_0_2, TRANS_PROB_0_3, TRANS_PROB_0_4, TRANS_PROB_0_5, TRANS_PROB_0_6, TRANS_PROB_1_0, TRANS_PROB_1_1, TRANS_PROB_1_2, TRANS_PROB_1_3, TRANS_PROB_1_4, TRANS_PROB_1_5, TRANS_PROB_1_6, TRANS_PROB_2_0, TRANS_PROB_2_1, TRANS_PROB_2_2, TRANS_PROB_2_3, TRANS_PROB_2_4, TRANS_PROB_2_5, TRANS_PROB_2_6, TRANS_PROB_3_0, TRANS_PROB_3_1, TRANS_PROB_3_2, TRANS_PROB_3_3, TRANS_PROB_3_4, TRANS_PROB_3_5, TRANS_PROB_3_6, TRANS_PROB_4_0, TRANS_PROB_4_1, TRANS_PROB_4_2, TRANS_PROB_4_3, TRANS_PROB_4_4, TRANS_PROB_4_5, TRANS_PROB_4_6, TRANS_PROB_5_0, TRANS_PROB_5_1, TRANS_PROB_5_2, TRANS_PROB_5_3, TRANS_PROB_5_4, TRANS_PROB_5_5, TRANS_PROB_5_6, TRANS_PROB_6_0, TRANS_PROB_6_1, TRANS_PROB_6_2, TRANS_PROB_6_3, TRANS_PROB_6_4, TRANS_PROB_6_5, PERC_STEPS_UNFINISHED, PERC_HELP_STEP, PERC_HELP_H0, PERC_HELP_H1, PERC_HELP_H2, PERC_HELP_H3, PERC_INCORRECT
# The activity model attributes to use when clustering that are nominal
# (mandatory)
attributes.nominal = STUDENT,PROBLEM, COMPLETED
# The weights for student and problem entropy, variance and prediction error
# (optional, value of each defaults to 1.0 if not specified)
weight.student_entropy = 1.0
weight.problem_entropy = 1.0
weight.standard_deviation = 1.0
weight.prediction_error = 1.0
# Whether detailed output for the clustering should be included in the results' file
# (optional, default is true)
detailed_output = true
#=============================================================================
References
Koedinger, K.R., Baker, R.S.J.d., Cunningham, K., Skogsholm, A., Leber, B. and Stamper, J.: A Data Repository for the EDM community: The PSLC DataShop. Handbook of Educational Data Mining, 2010.
VanLehn, K., Lynch, C., Schulze, K., Shapiro, J.A., Shelby, R.H., Taylor, L., Treacy, D., Weinstein, A., and Wintersgill, M.: The Andes Physics Tutoring System: Lessons Learned. International Journal of Artificial Intelligence in Education 15(3), 2005.
Köck, M., and Paramythis, A.: Activity Sequence Modelling and Dynamic Clustering for Personalized E-Learning. User Modeling and User-Adapted Interaction, 2011.
